Hormone-producing cells in the human gut are difficult to study, as they are very rare and unique to different species of animals. Researchers from the Hubrecht Institute and Utrecht University developed an extensive toolbox to study human hormone-producing cells in tiny versions of the gut grown in the lab, called organoids. Organoids allowed them to uncover secrets of the human gut, for example which potential hormones can be made by the gut and how the secretion of these hormones is triggered. These findings offer potential new avenues for the treatment of diseases such as Type 2 diabetes and obesity.
The gut hormone challenge
Enteroendocrine cells only comprise about 1% of the lining of the gut. To complicate this further, more than 20 different hormones are produced by at least 5 subtypes of these enteroendocrine cells of which some are even more rare. This would mean you would have to look at more than 1000 cells to find an enteroendocrine cell that makes a specific hormone.
A human gut hormone toolbox
Researchers from the group of Hans Clevers at the Hubrecht Institute overcame this issue, in a creative way. They used intestinal organoids, tiny intestines that they can grow in the lab from human stem cells, and increased the activity of a gene called neurogenin-3. This dramatically increased the number of enteroendocrine cells in these mini-intestines, while still generating all the different subtypes of these cells.
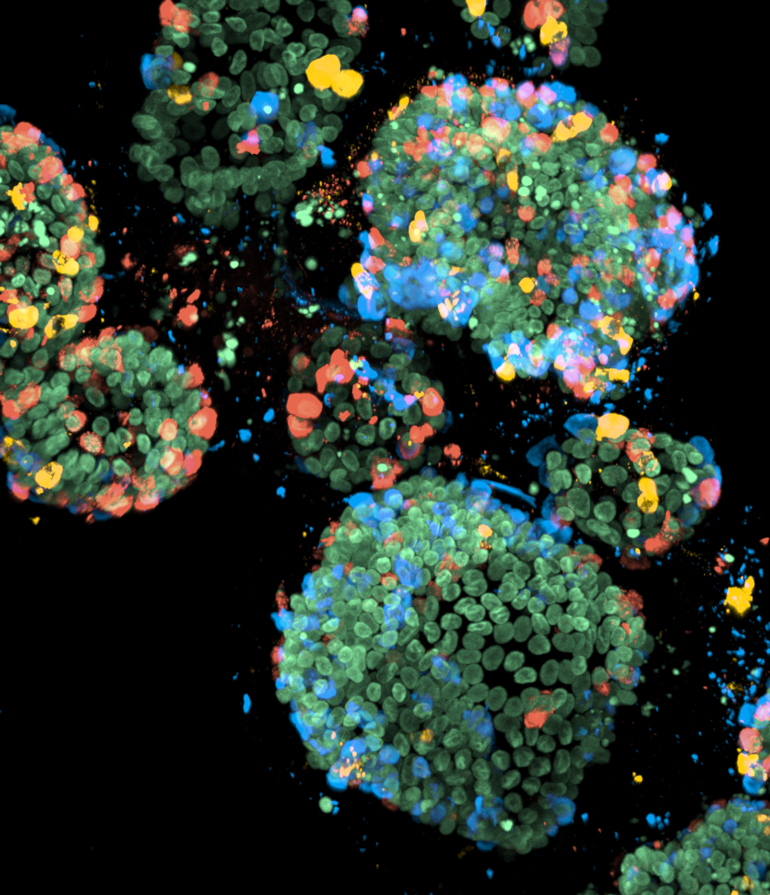
To be able to study all the specific types of enteroendocrine cells, the researchers used another trick that was recently developed in the group of Hans Clevers. Clevers: “In our lab, we have optimized genetic engineering of organoids. We were therefore able to label the hormones that are made by the enteroendocrine cells in different colors and create a biobank of mini-intestines, called the EEC-Tag biobank, in which different hormones are tagged with different colors.” When an enteroendocrine cell starts producing a labeled hormone, that cell will appear in the corresponding color. The researchers can use the EEC-Tag biobank to study ten major hormones and different combinations of these hormones within the same organoid.
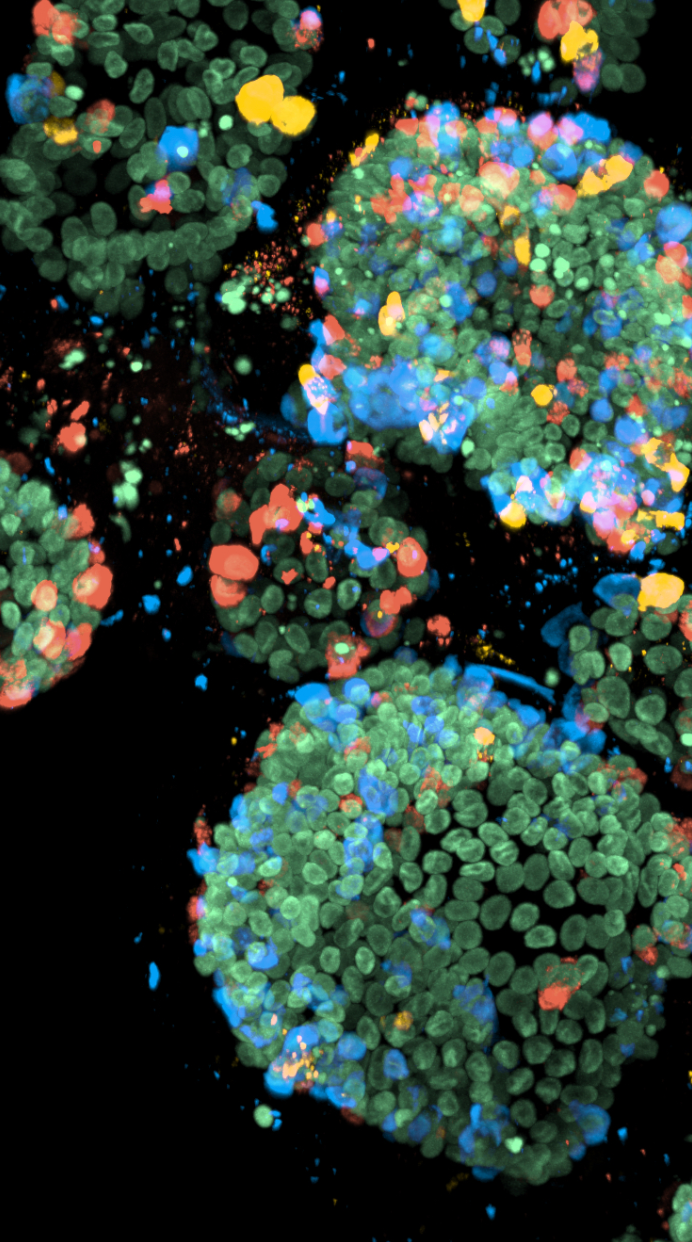
New molecules
For the first time, the researchers described all the genes that are active in the different types of human enteroendocrine cells, using a technique called single-cell sequencing. Based on these data they created an atlas with the deepest insights to date into the different subtypes of human enteroendocrine cells. The atlas revealed the activity of genes pivotal to the function of enteroendocrine cells that were not described before. For example, new receptors were identified that these cells can use to sense food and release their hormones. Other discoveries include potential hormones that have never been identified in the gut before.
Large scale screens
With all these data and tools at hand, researchers can now do large scale screens to study which molecules in our food are sensed by which types of enteroendocrine cells, how the enteroendocrine cells actually sense these molecules, and which hormones are produced as a response. Clevers: “The combined ability to make large numbers of all human enteroendocrine cells and track the subtypes using fluorescent colors in organoids makes it possible to perform screens for drug targets that may harness the therapeutic potential of enteroendocrine cells”. This work paves the way for further investigation of the largest hormone-producing organ in our body and may eventually lead to treatments for people affected by obesity and diabetes.
Publication
High Resolution mRNA and Secretome Atlas of Human Enteroendocrine Cells. Joep Beumer*, Jens Puschhof*, Julia Bauzá-Martinez*, Adriana Martínez-Silgado, Rasa Elmentaite, Kylie R. James, Alexander Ross, Delilah Hendriks, Benedetta Artegiani, Georg Busslinger, Bas Ponsioen, Amanda Andersson-Rolf, Aurelia Saftien, Charelle Boot, Kai Kretzschmar, Maarten H. Geurts, Yotam E. Bar-Ephraim, Cayetano Pleguezuelos Manzano, Yorick Post, Harry Begthel, Franka van der Linden, Carmen Lopez-Iglesias, Willine J. van de Wetering, Reinier van der Linden, Peter J. Peters, Albert .J.R. Heck, Joachim Goedhart, Hugo Snippert, Matthias Zilbauer, Sarah A. Teichmann, Wei Wu#, Hans Clevers#. Cell 2020.
* these authors contributed equally, # senior researchers




